Our RNA backbone research has now been updated from the earlier work reported in: Murray et al (2003), "RNA backbone is rotameric", PNAS 100: 13904-13909. The new system (Richardson et al (2008), "RNA backbone: Consensus all-angle conformers and modular string nomenclature" doi/10.1261/rna.657708 publication submitted) is a community-consensus effort under the auspices of the RNA Ontology Consortium. It defines a new modular nomenclature for RNA full-detail backbone conformation, and reports an updated and expanded consensus list of backbone rotamers - 46 official conformers, plus another 7 "wannabes". These are now built into the MolProbity analyses of RNA structures and can also be assigned by the Suitename program available in our software section. This description on this page, below, refers to the older system. The new conformers, with graphics for a representative example of each, are described on this table [note that it takes a few seconds to load completely].
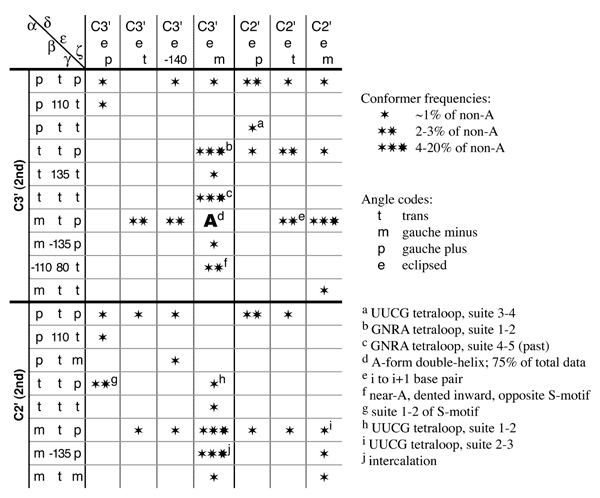
Nucleic acid backbone is important in many processes, from participating directly or positioning bases at ribozymes' catalytic sites to engaging in specific binding to proteins or smaller molecules. It is still a challenging part of molecular structures to determine: there are at least six degrees of freedom to be considered for each residue, and often the data is not entirely sufficient. In X-ray crystallography, the bases and phosphates can be distinguished well, but the regions in between are variable and the density is often ambiguous for at least part of them. In 1H NMR, the bases and sugars can be determined fairly well, but there aren't enough other constraints. It turns out, however, that there is another natural division into base-to-base units -- or, from a backbone perspective, slightly overlapping two-sugar units, dubbed "suites" -- on the grounds that this is the unit within which the most interactions along the chain occur. Within a suite, the available combinations of dihedral values are more restricted.