When there is a need to divide up protein structure for purposes of description, prediction, spectroscopic characterization, etc., the usual categories have been "helix," "β structure," and "coil." "Coil" is defined in practice as "none of the above." Its major distinguishing feature is that it is nonrepetitive in backbone conformation (although depending on one's definition of β structure, one might or might not include an isolated piece of extended chain as coil). Sometimes coil is referred to as "random coil," or is modeled by properties observed in 6M guanidine hydrochloride. This seems unfortunate, since the actual portions of crystallographically determined protein structures generally described as coil are not random or disordered in any sense of the word; they are every bit as highly organized and firmly held in place as the repeating secondary structures — they are simply harder to describe. In recent years since recognition of the wide occurrence and importance of tight turns (see Section II,C), they are often separated out as a category of structure, so that now the miscellaneous "coil" category would refer to what is neither helix, β, nor turn.
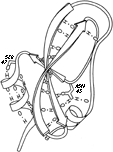
FIG. 53. The main chain hydrogen bonds of basic pancreatic trypsin inhibitor, plus two of the side chains whose hydrogen bonds stabilize the ends of pieces of secondary structure: Ser-47 at the beginning of an α-helix and Asn-43 at the end of a β strand.
Turns and bulges are nonrepetitive features which are characterized primarily in terms of backbone conformation and backbone hydrogen bonding. However, much coil structure appears to be very strongly influenced by specific side chain interactions. These have not been very widely analyzed, but a few examples can illustrate the sorts of patterns to be expected. Probably the earliest notice of such a feature is the observation in Kendrew et al. () that serine or threonine frequently hydrogen-bonds to a backbone NH exposed at the beginning of an α-helix. Figure 53 illustrates such a conformation. [This is now called a helix N-cap (Richardson, 1988) and is discussed in section II, A.] Energy calculations for side chain interactions have, quite understandably, considered only the very local region (e.g., ). Recently there have been some systematic empirical computer surveys of side chain environments, such as in Warme and Morgan () and Crippen and Kuntz (), and surveys of side chain conformations, such as Janin et al. () and Bhat et al. (). These do not focus specifically on nonrepetitive structure, but any strong preferences would be especially influential there. So far, however, these studies are still at the initial stages of tabulating raw statistical preferences. [Later work has tabulated the preferred conformations of side-chain "rotamers" (e.g., Lovell et al., 2000), including the preferred patterns of local sidechain-backbone H-bonds.] (ref ??)
Locations of the Asparagine Residues Are Tabulated for β Sheets of at Least Three Strands (and Known Amino Acid Sequence) in the Known Protein Structuresa | |||
Asparagine position in β sheets | |||
At end of strand |
On a side strand |
In middle of sheet | |
Antiparallel |
30 |
17 |
1 |
Parallel |
10 |
4 |
3 |
a For all residues in β sheet, approximately 50% are in the middle and 50% at either an end or a side. In contrast, less than 20% of the asparagines in parallel sheet are in the middle, and only 2% of those in antiparallel sheet are in the middle. | |||
At the other extreme, it is possible to examine individual examples of a single potentially interesting type to see simply what features turn up. Asparagine is one such potentially interesting residue, since it combines a side chain that mimics a backbone peptide, along with the conformational constraints of possessing only two side chain variable angles. Asn is more likely than any other non-glycine residue to have φ,ψ angles outside the normally allowed regions. Also, it does indeed show a pattern of hydrogen-bonding that is distinct from either glutamine or aspartate. Asparagine is more than twice as likely as any other residue to bond to the first exposed backbone NH or CO at the end of an antiparallel β strand (as in, for example, Fig. 53) and probably for that reason is essentially forbidden from occurring in the interior of antiparallel β sheet although it is very common at or just beyond the edges (see Table I). Asparagine also shows a favored type of hydrogen bond from its side chain CO to the backbone NH of residue n + 2. This pattern happens to predispose a type I tight turn at residues n + 1 and n + 2 (see Fig. 54). [This Asn H-bond also itself mimics a tight turn and has been called a pseudo-turn (Ress)] (ref ??); such cases are treated in detail by ?? ref.
FIG. 54. An asparagine side chain making a hydrogen-bond to the main chain NH of residue n + 2, an arrangement which helps stabilize the central peptide of a tight turn. Residues 91-93 from chymotrypsin.
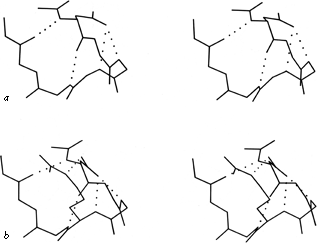
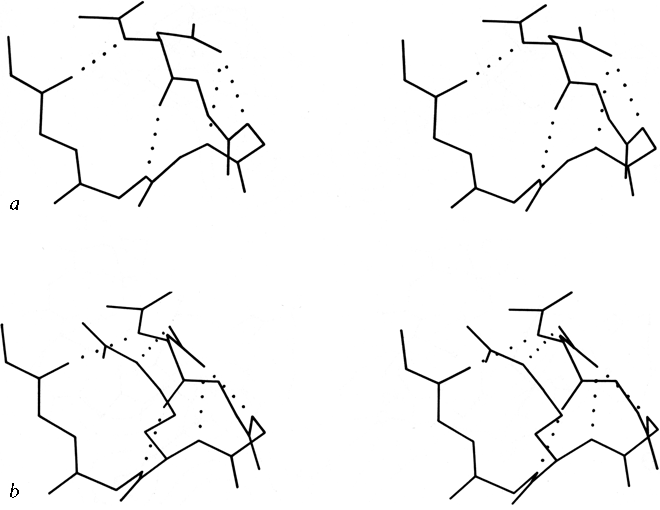
FIG. 55. Two very similar 5-residue turns with a single α-helical hydrogen bond: (a) pancreatic trypsin inhibitor residues 24-28, stabilized by the side chain of Asn-24; (b) prealbumin residues 18-22, stabilized by Asp-18.
Figure 55a shows a small piece of nonrepetitive structure from pancreatic trypsin inhibitor in which the side chain 0δ of Asn-24 is hydrogen-bonded to both the n + 2 and n + 3 backbone NH groups. There is a G1 β bulge (see Section II,D) at Gly-28, Leu-29, Asn-24. The corner between the two strands is turned by what could be better described as a five-residue turn with one α-helical hydrogen bond and an Asn stabilizing the loop NH groups, rather than as two successive non-hydrogen-bonded turns (which is how it shows up in any computer search for tight turns). An extremely similar conformation occurs in prealbumin (), with the same G1 bulge and five-residue "α" turn stabilized this time by Asp-18 instead of an asparagine, and with an additional bond to the Nε of Arg-21 (see Fig. 55b).
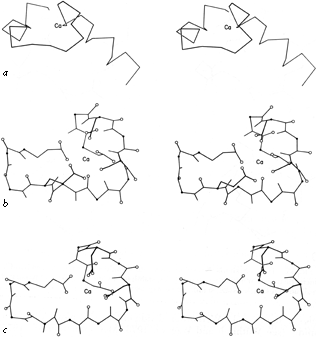
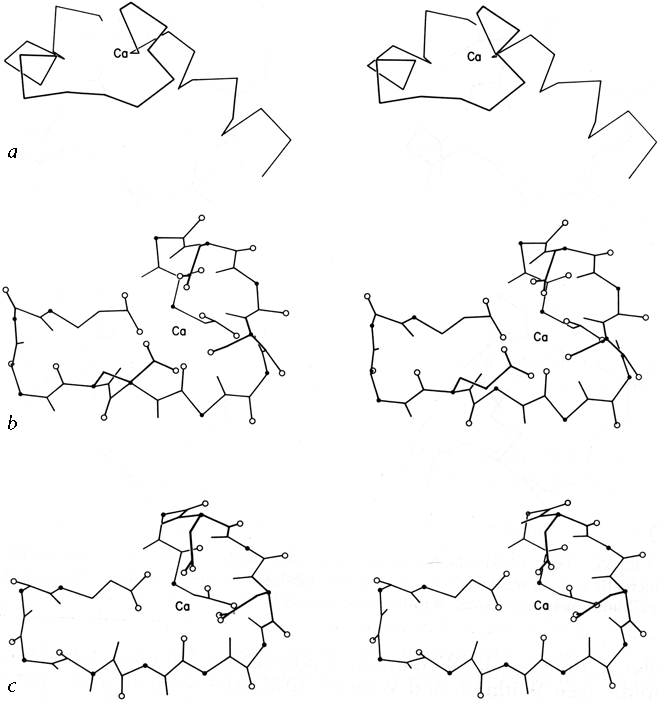
FIG.56. The calcium-binding sites from carp muscle calcium-binding protein: (a' backbone of the entire "E-F hand"; (b) detailed view of the E-F calcium-binding site, including those side chains which are Ca ligands; (c) detailed view of the C-D calcium-binding site, rotated to match part b. Oxygens are shown as open circles and α-carbons as solid dots.
It will be interesting in the future to see fuller characterization of nonrepetitive structure, especially since it forms many of the more complicated enzyme active sites. A few instances have so far been described in which a particular organization of coil structure can be recognized as providing a particular type of functional site, since it occurs with very similar patterns in more than one example. One of these structures is a loop which binds iron-sulfur clusters in both ferredoxin and high-potential iron protein (). Another such structure is the central loop portion of the "E-F hands" that form the calcium-binding sites in carp calcium-binding protein (). The backbone structures curl around in very similar conformations and provide ligands in a definite order to the six octahedral coordination sites around the Ca(2+), as illustrated in Fig. 56.