In contrast to the well-ordered but nonrepetitive coil structures, there are also genuinely disordered regions in proteins, which are either entirely absent on electron density maps or which appear with a much lower and more spread out density than the rest of the protein. The disorder could either be caused by actual motion, on a time scale of anything shorter than about a day, or it could be caused by having multiple alternative conformations taken up by the different molecules in the crystal. Well-ordered side chains also move, often very rapidly (see ; ), but the movements are brief departures from a single stable conformation.
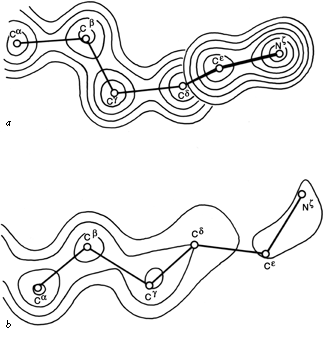
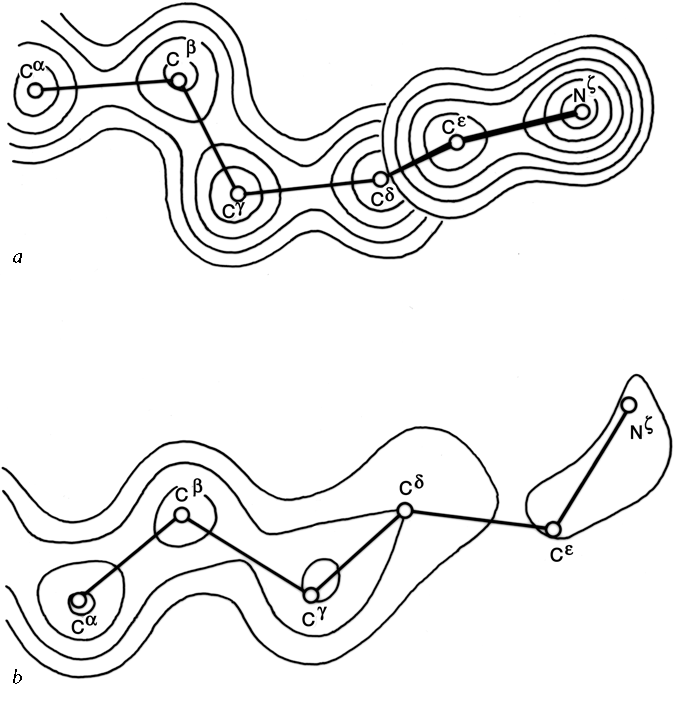
FIG.57. Model and electron density in rubredoxin after refinement at 1.2Å resolution, for (a) the well-ordered lysine, Lys-46 (temperature factor average of 9.2 for the outer four atoms of the side chain); (b) the best of the disordered lysines, Lys-3 (temperature factor average of 43.6 for the outer four side chain atoms). From Watenpaugh et al. (), Fig.12, with permission.
One simple case of disordered structure involves many of the long charged side chains exposed to solvent, particularly lysines. For example, 16 of the 19 lysines in myoglobin are listed as uncertain past Cδ and 5 of them for all atoms past Cβ (); for ribonuclease S Wyckoff et al. () report 6 of the 10 lysine side chains in zero electron density; in trypsin the ends of 9 of the 13 lysines refined to the maximum allowed temperature factor of 40 (R. Stroud and J. Chambers, personal communication); and in rubredoxin refined at 1.2 Å resolution the average temperature factor for the last 4 atoms in the side chain is 9.2 for one of the four lysines versus 43.6, 74.4, and 79.3 for the others. Figure 57 shows the refined electron density for the well-ordered lysine and for the best of the disordered ones in rubredoxin. Interestingly, arginine side chains do not follow this same pattern. In the structures quoted above, 70% of the arginines were well ordered, as opposed to only 26% of the lysines. In refinement at very high resolution it is sometimes possible to express a partially disordered side chain as a mixture of two different specific conformations, as, for instance, isoleucines 7 and 25 in crambin (). [Alternate conformations have become much more routinely identified now that more structures are solved at very high resolutions. In crambin at 0.54 Å resolution (1EJG), many sidechains, and even parts of the backbone, have alternate conformations, while some sidechains are clearly seen in three different conformations.]
It is fairly common for a few residues at the N-terminus or the C-terminus of the polypeptide chain to be disordered, if they include no large hydrophobic residues. In some cases it is known that these dangling ends can be cleaved off without any loss of stability or activity in the protein (e.g., ). However, there are certainly other cases in which disordered regions have very definite functional roles. Sometimes there is disorder in one state and an ordered conformation in another state, with the contrast between the two having a functional role; for example, the inter-subunit salt linkages in hemoglobin which provide constraints in the deoxy form and are free and disordered in the oxy- form (, ). In systems designed for specific proteolytic cleavage, disorder is one way of promoting cleavage at a given loop. The new chain ends liberated by such cleavage are also very often disordered, as for instance in chymotrypsin (). In some cases a ligand-binding site may show partial or complete disorder in the absence of the ligand but become well-ordered when the ligand is bound, as for instance the RNA site of tobacco mosaic virus protein (). [In triose phosphate isomerase a partially disordered loop over the active site region has, surprisingly, a highly conserved sequence. It turns out this loop becomes ordered on substrate binding, protecting the reaction from solvent and cleverly trading off a loss of entropy against a gain in binding enthalpy to give high specificity without a very tight binding constant that would hinder product release (Petsko).]
One of the most intriguing recent examples of disordered structure is in tomato bushy stunt virus (), where at least 33 N-terminal residues from subunit types A and B, and probably an additional 50 or 60 N-terminal residues from all three subunit types (as judged from the molecular weight), project into the central cavity of the virus particle and are completely invisible in the electron density map, as is the RNA inside. Neutron scattering () shows an inner shell of protein separated from the main coat by a 30-Å shell containing mainly RNA. The most likely presumption is that the N-terminal arms interact with the RNA, probably in a quite definite local conformation, but that they are flexibly hinged and can take up many different orientations relative to the 180 subunits forming the outer shell of the virus particle. The disorder of the arms is a necessary condition for their specific interaction with the RNA, which cannot pack with the icosahedral symmetry of the protein coat subunits.
Although disordered structure is fairly common in the known protein structures, this is undoubtedly one of the cases in which the process of crystallization induces a bias on the results observed. Since extensive disorder makes crystals much harder to obtain, it seems probable that disordered regions are even more prevalent on the proteins that do not crystallize.