The largest grouping of structures contains domains organized around a parallel or mixed β sheet, the connections for which form structure (usually helical) protecting both sides of the sheet, with the helices also predominantly parallel to each other. Of course, each helix and its neighboring β strand are antiparallel to one another, but this structure category is called parallel α/β because both the β sheet interactions and the α-helix interactions are internally parallel. The parallel α/β category is the same as Levitt and Chothia's α/β proteins. Figures 75-78 show schematic drawings of this group of structures. It is interesting to note that there seems no a priori reason not to have parallel all-α structures or parallel all-β structures formed of two helix layers or of two parallel β sheets, yet such structures are not found. [There are now indeed examples of both these types, described in the annotations to sections III.B for helical horseshoes and III.D. for β-helices.] All of the domains with parallel organization have both a β sheet of at least four or five strands and at least three or four α-helices. Almost all have at least three layers.
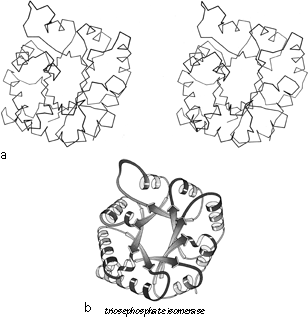
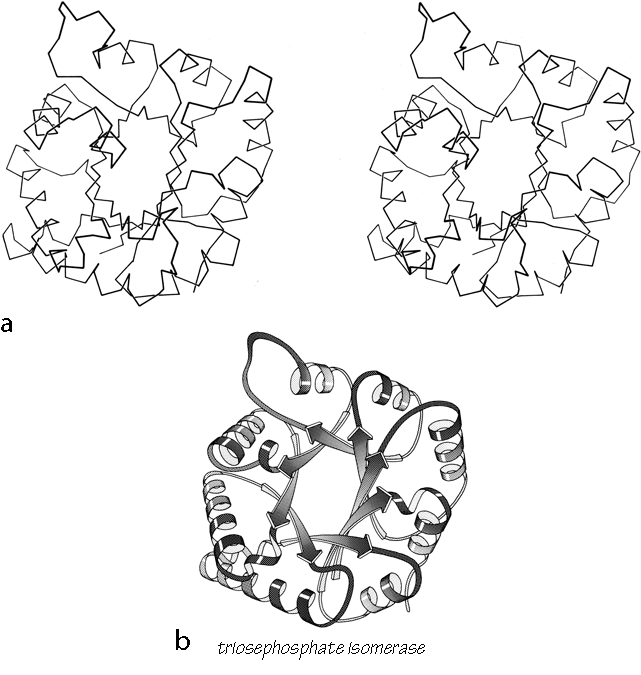
FIG. 90. Triosephosphate isomerase as an example of a singly wound parallel β barrel. (a) α-Carbon stereo, viewed from one end of the barrel; (b) backbone schematic, viewed as in a; (c) α-carbon stereo, viewed from the side of the barrel; (d) backbone schematic, viewed as in c; (e) topology diagram showing the +1x right-handed connections between the β strands.
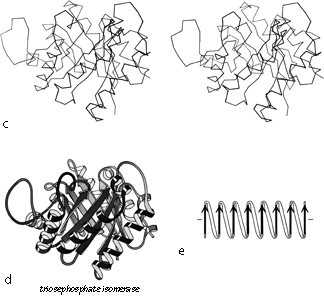
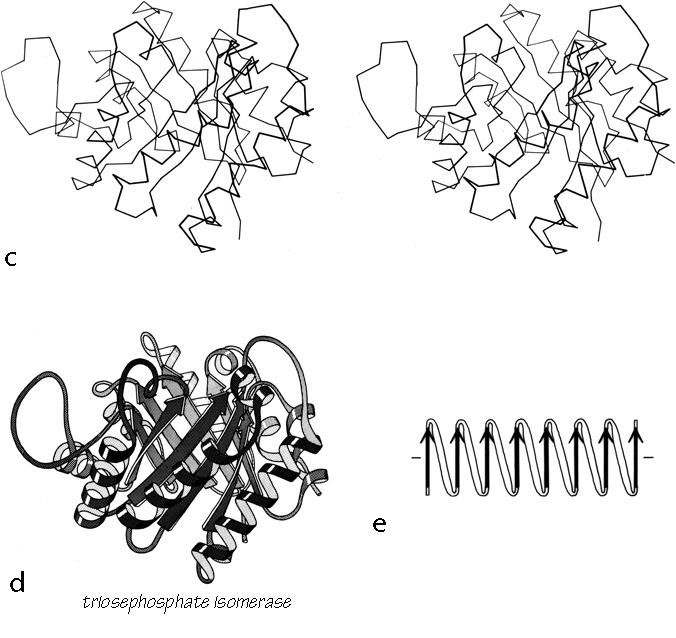
FIG. 90 (continued)
The first subgrouping under the parallel α/β category contains two of the largest but simplest domain structures that have yet been found. They are the eight-stranded parallel β barrels of triosephosphate isomerase (see Fig. 90) and pyruvate kinase domain d1, both of which are connected in +1x,+1x topology all the way around. (In structures with both β sheet and also several helices it is convenient to use just the β strands for designating the topology.) The connections are α-helices, which form a larger cylinder of parallel helices concentric with the β barrel. The structural elements of both α and β cylinders have a pronounced right-handed twist around the cylinder axis. Connections between the parallel β strands must lie on the outside of the barrel since the interior is filled by the packed hydrophobic side chains. If all of the crossover connections must be right-handed and no knots are allowed, then the chain must wind consistently around the barrel in one direction, and the +1x, +1x, +1x topology is not only the simplest but essentially the only possible topology for such a structure (), since all other topologies are knotted and unfoldable. We call this structure the singly wound parallel β barrel, since successive crossover connections are wound on the barrel progressing in a single direction with no reversal or backtracking. Figure 91a is a highly schematized representation of the "singly wound" structure, viewed from one end of the barrel.
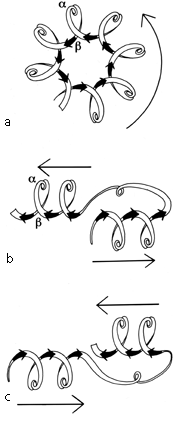
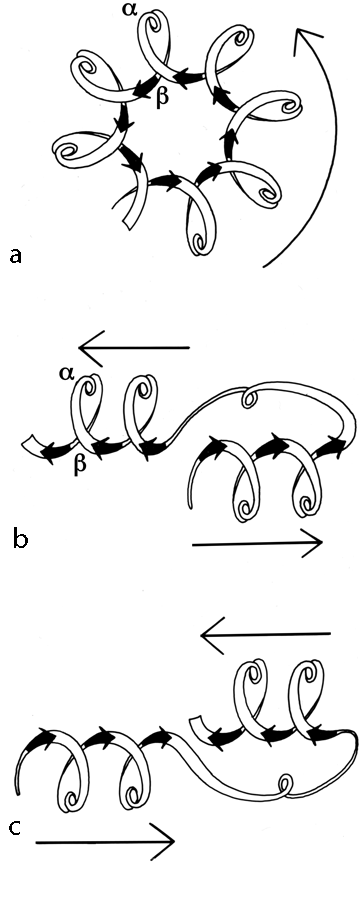
FIG. 91. Highly simplified sketches (viewed from the C-terminal end of the β strands) of (a) a singly wound parallel β barrel; (b) a classic doubly wound β sheet; (c) a reverse doubly wound β sheet. Thin arrows next to the diagrams show the direction in which the chain is progressing from strand to strand in the sheet.
The largest subgrouping within the parallel α/β category contains structures with a central twisted wall of parallel or mixed β sheet, protected on both sides by its crossover connections (most of which are helical). This is called the doubly wound parallel β sheet, because with right-handed crossovers the simplest way of protecting both sides of the sheet is to start near the middle and wind toward one edge, then return to the middle and wind to the other edge. Figure 91b is a highly schematized representation of the "doubly wound" structure, viewed from one end of the sheet (compare with Fig. 91a). The singly wound barrel has four major layers of backbone structure and the doubly wound sheet has three major layers (with two separate hydrophobic cores); most other domain structures have only two major backbone layers with a single hydrophobic core, and are on the average considerably smaller.
The doubly wound structures were first recognized as a category by Rossmann in comparing flavodoxin with lactate dehydrogenase d1. As more and more protein structures were solved which fell into this category, the relationships between them have been described and debated at considerable length. The initial descriptions were in terms of the β—α—β—α—β unit as a supersecondary structure (). Quite soon the emphasis shifted to the functional properties of the nucleotide-binding site which most of them share, and to the probable evolutionary relationships between these "nucleotide-binding domains" (; ). By now the consensus appears to be that some of the most similar of these structures must certainly be related to each other, while at least some of the most dissimilar examples surely cannot be related (; ; ; ).
We will group these domains into five gradually loosening levels of topological similarity, without attempting to make any definite decision as to where the dividing line lies between divergent and convergent examples.
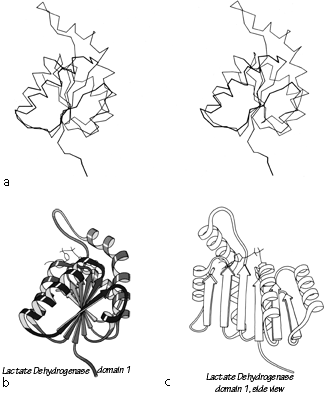
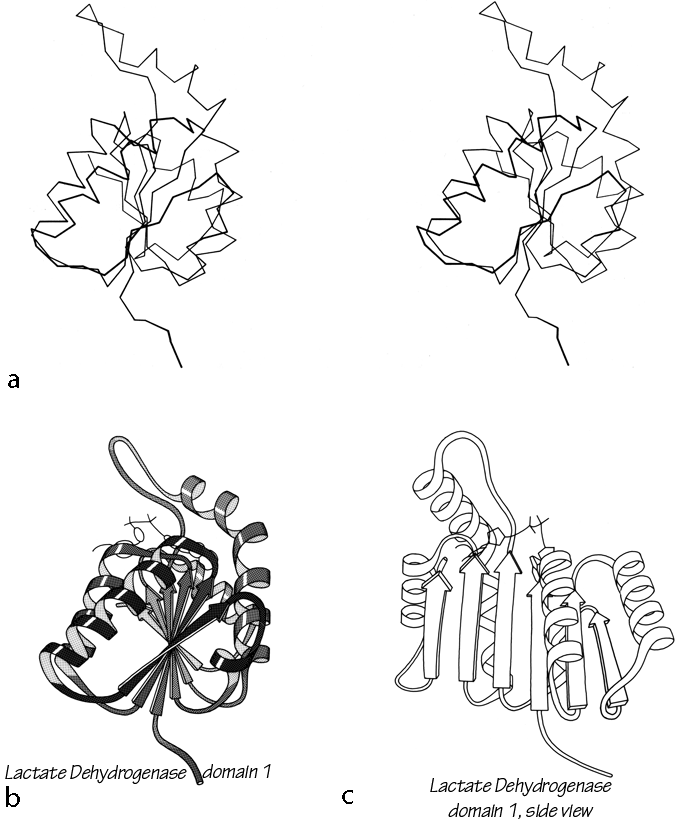
FIG. 92. Lactate dehydrogenase domain 1 as an example of a classic doubly wound parallel β sheet. (a) α-Carbon stereo, viewed from one edge of the sheet; (b) backbone schematic, viewed as in a; (c) backbone schematic, viewed from one face of the sheet.
Figure 92 shows stereo and schematic drawings of lactate dehydrogenase domain 1, which is the classic example of a nucleotide binding domain or doubly wound sheet.
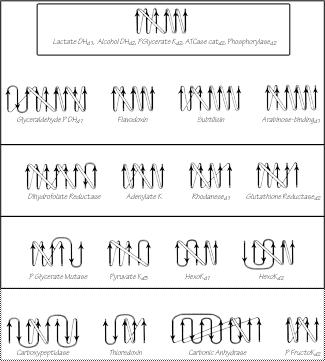
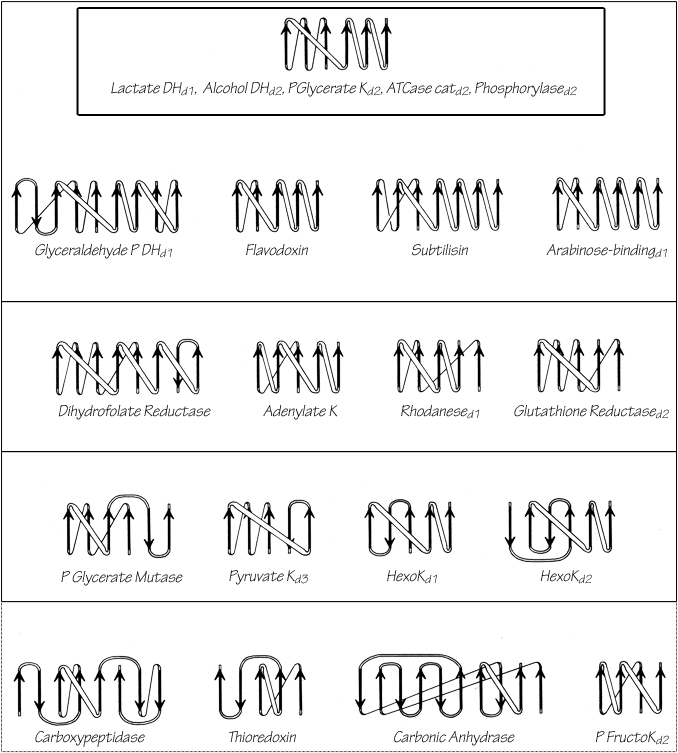
FIG. 93. Topology diagrams for the doubly wound and miscellaneous α/β domains illustrated in Figs. 76 through 78. Arrows represent the β strands; thin connections lie behind the β sheet and fat ones above it. The darkest upper box surrounds the classic doubly wound sheets; successively lighter solid boxes include domains that are progressively less like the classic topology; the dotted box encloses the miscellaneous α/β structures. K = kinase; P = phospho; DH = dehydrogenase; ATCase = aspartate transcarbamylase.
The darkest inner box in Fig. 93 includes those "classic" doubly wound parallel sheets that exactly match the topology of lactate dehydrogenase d1. Phosphorylase domain 2 is a five-layer structure in which the central three layers are a classic doubly wound sheet and the outer helical layers are formed by the two ends of the chain. The next box includes examples in which deleting one or two strands either at an end of the chain or at an edge of the β sheet will produce a five- or six-strand doubly wound sheet, while in the next box such deletions yield four doubly wound strands. In the outermost solid box it is necessary to omit strands interior to both the sheet and the sequence in order to get four strands of doubly wound β sheet. The structures inside the dotted box can yield no more than three such strands and cannot really be described as doubly wound, they share with the rest of this large subgrouping only the general organization of a central wall of parallel or mixed β sheet protected on both sides by its connections (see Fig. 94 for carboxypeptidase as an example). As one progresses outward from the classic to the most peripheral cases, the number of antiparallel strand pairs mixed in with the parallel gradually increases. Aside from the "classic" examples in the inner box, there are several other exact duplicates of doubly wound topologies between different proteins: phosphorylase d1 versus glyceraldehyde-phosphate dehydrogenase d1; aspartate transcarbamylase catalytic d1 versus rhodanese d1,d2; catalase d3 versus flavodoxin; and ρ-hydroxybenzoate hydroxylase d1 versus glutathione reductase d1.
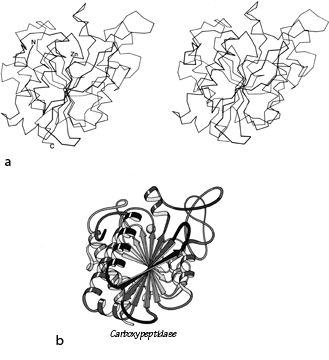
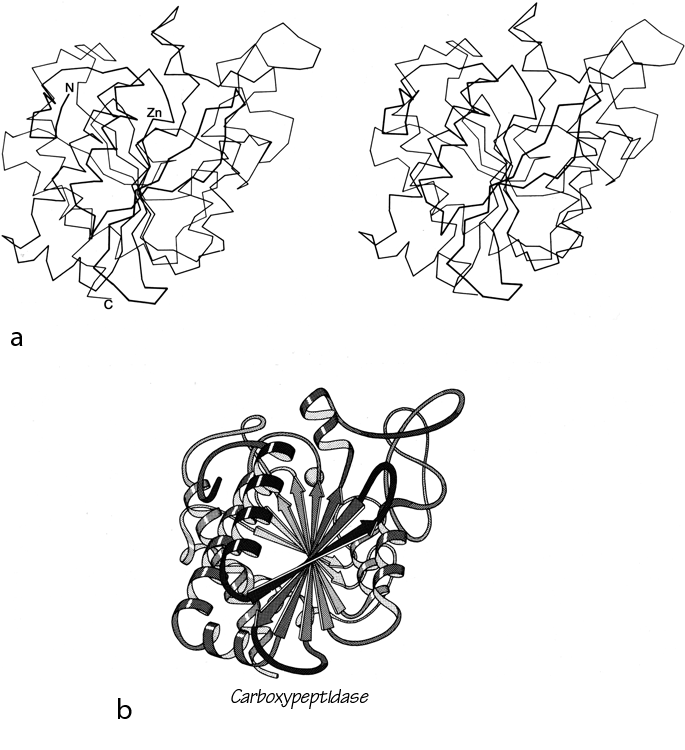
FIG. 94. Carboxypeptidase A as an example of a miscellaneous α/β structure. (a) α-Carbon stereo, viewed from one edge of the mixed β sheet; (b) backbone schematic, viewed as in a.
As one progresses from classic to peripheral doubly wound sheets, the number of domains that bind nucleotides also decreases. A favorable site for binding dinucleotides (or in a few cases, mononucleotides) is associated with this general category of structure and to a large extent with the doubly wound topology. The dinucleotides are all bound in approximately equivalent positions at the C-terminal end of the β sheet strands, within one strand of the central position where the winding switches direction (see Fig. 91b). [The importance of these "topological switch points" for active sites in doubly-wound domains is well explained in Branden and Tooze (1991).] Nucleotides are also bound at the C-termini of β strands in the singly wound barrels. In most of these cases, each nucleotide is associated with a "mononucleotide-binding fold" of three β strands and two helices with +1x, +1x topology; combination of two of these folds around a local 2-fold axis produces the classic doubly wound sheet. In some cases, however (such as hexokinase or dihydrofolate reductase), the topology is quite significantly different. Also there seems to be another quite different type of nucleotide-binding site such as the active site in staphylococcal nuclease () or the AMP site in phosphorylase (); both of these sites rely mainly on arginines for binding the nucleotide phosphates.
One quite surprising and intriguing feature of this group of structures is that it contains extremely few examples of the "reverse doubly wound" topology (see Fig. 91c), a different but equally plausible pattern related to the doubly wound sheet by reversing the N- to C-terminal direction of the chain or by switching relative positions of the two halves of the β sheet. Of the four reverse examples (found in PGK d2, GPDH d1, glucosephosphate isomerase d1, and phosphorylase d1) none forms a nucleotide-binding site, all belong to a sheet that also has a normal doubly wound section, and none includes more than four strands. Those cases do demonstrate that such a topology is stable and can fold, but there must be some strong reason why it is so rare. Some simple explanations of this regularity would be either that most of the nucleotide-binding domains are related, or that they must fold strictly from the N-terminus, or that the requirements for forming a nucleotide-binding site are restrictive enough to constrain the usual doubly wound topology. None of these explanations is completely satisfying, however, because a number of domains are known that cannot fold strictly from the N-terminus, because the relative placement of features forming the nucleotide sites is only rather approximate (e.g., see ), and because the rearrangement necessary to produce a reverse doubly wound sheet seems much less drastic than many of the rearrangements that must be proposed if all of these proteins are related.